Benjamin Rowland
Chromosome biology
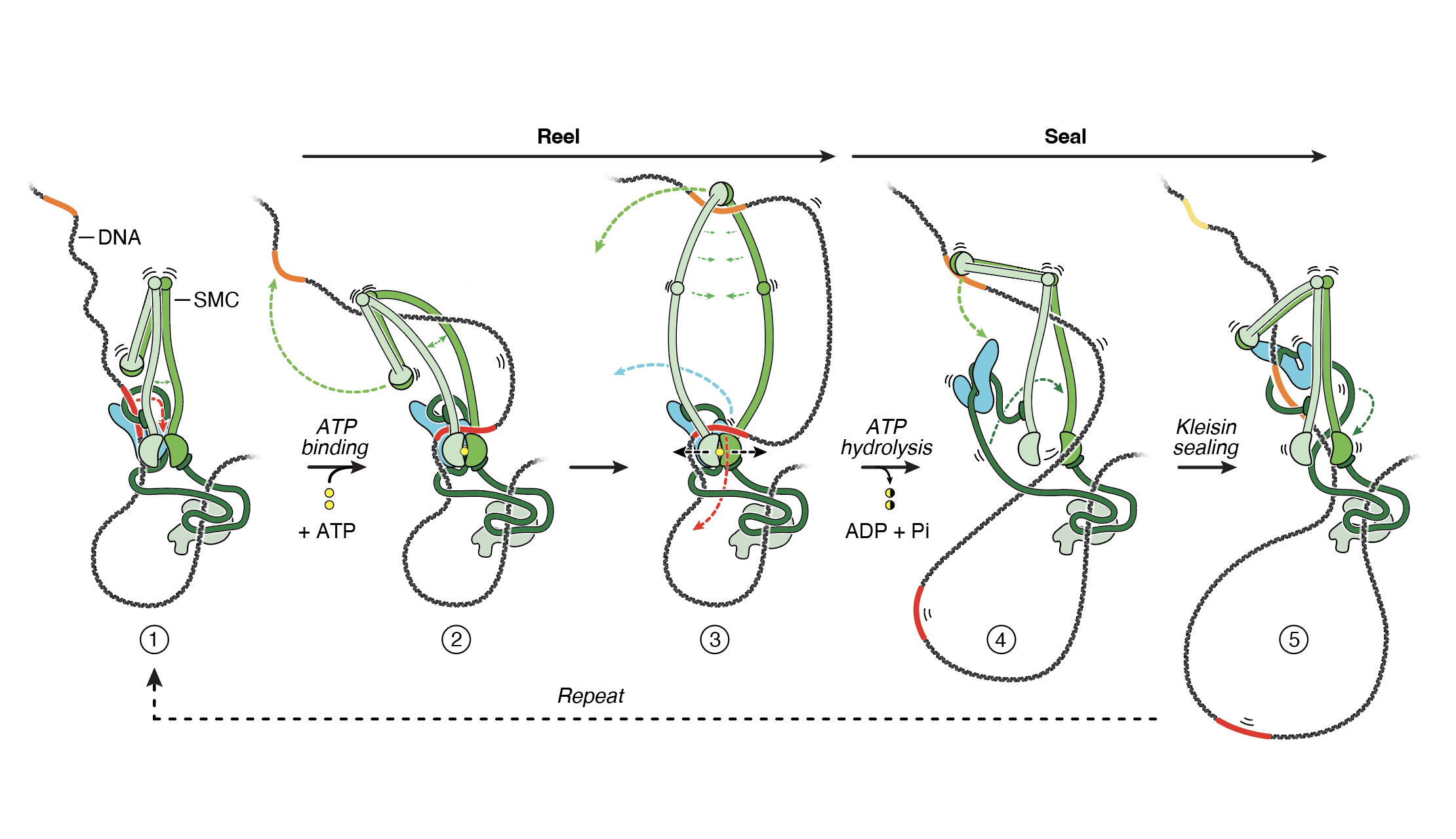
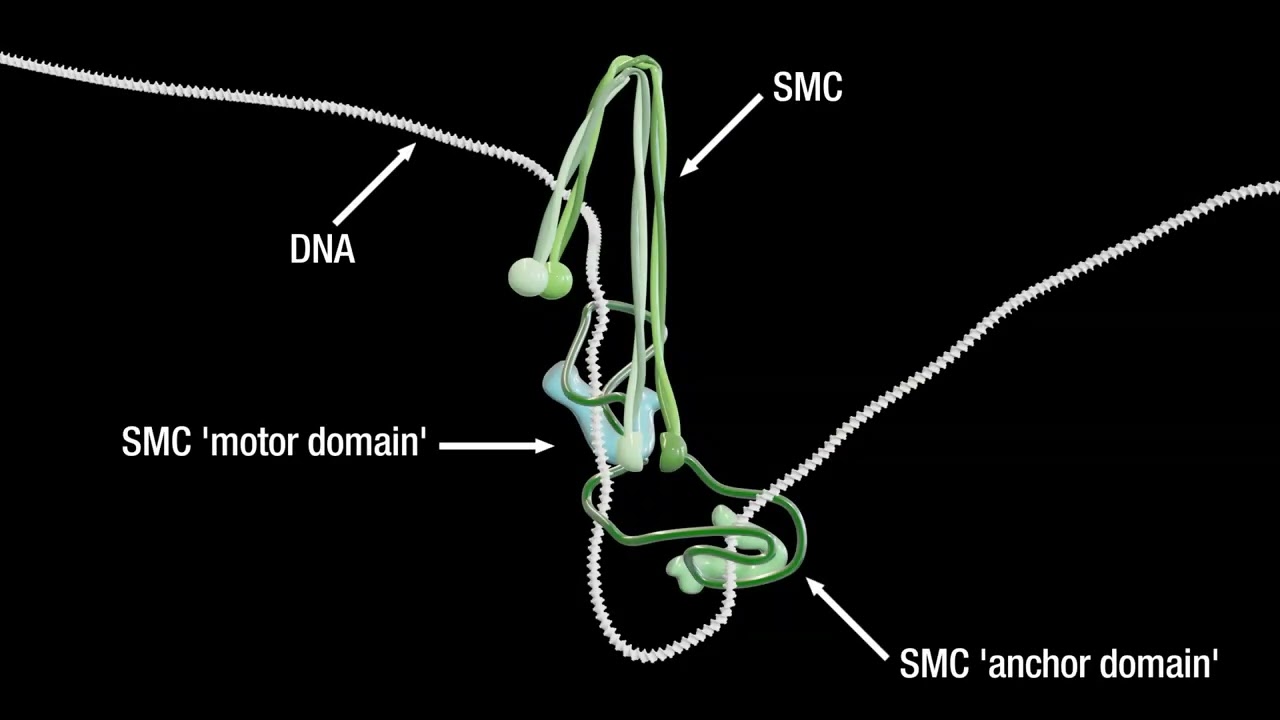
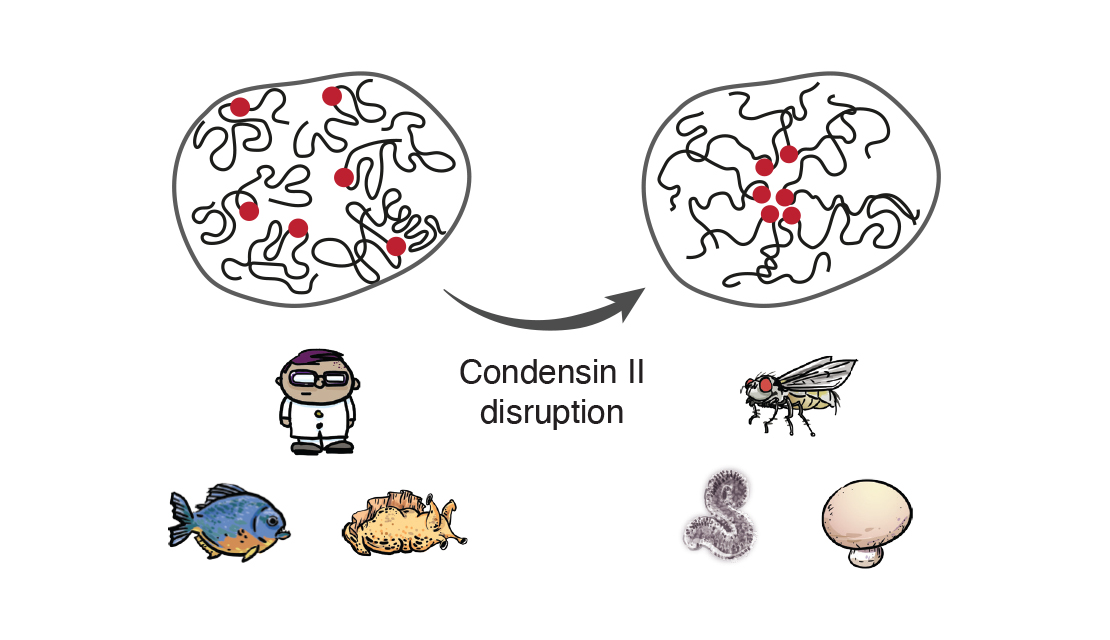
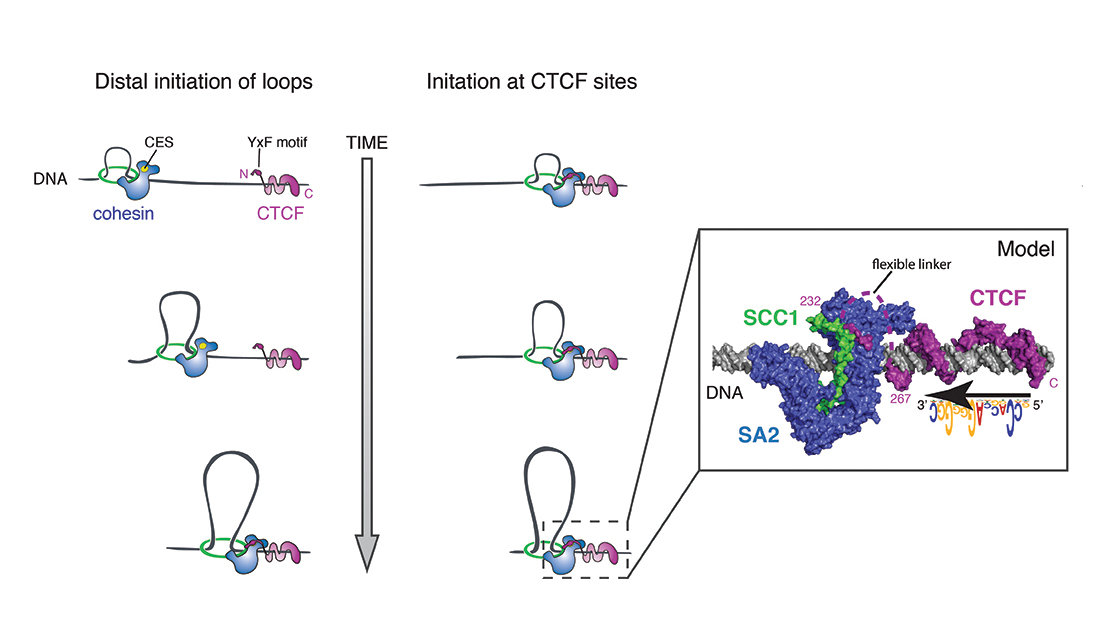
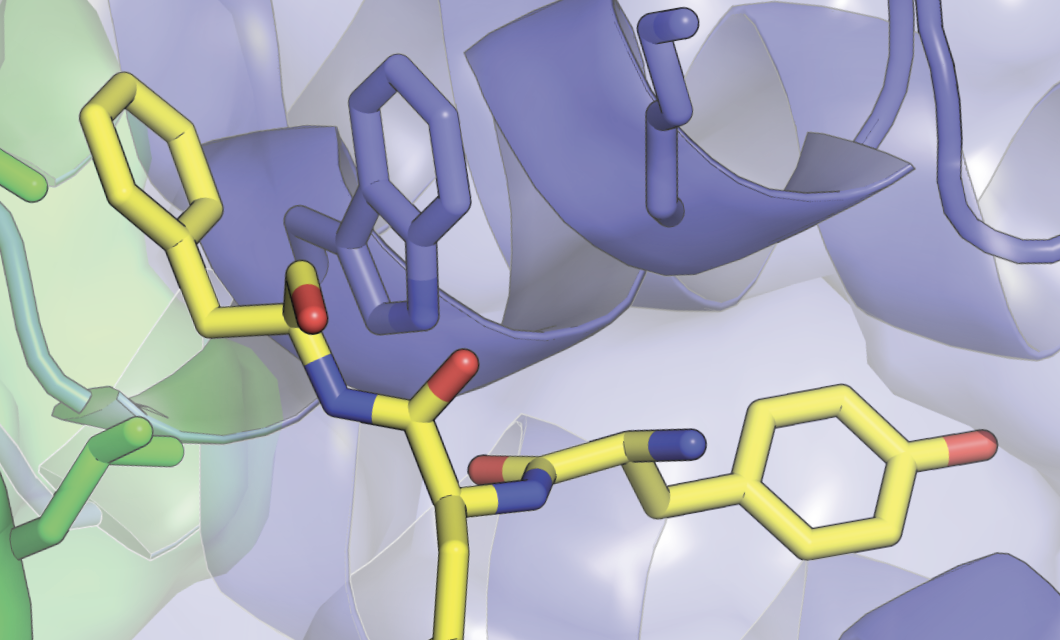
How do cohesin and condensin build DNA loops and shape the genome in 3D? How do these complexes entrap and release DNA? How does cohesin stably lock together the sister chromatids? How does condensin drive mitotic chromosome condensation? How does the SMC5/6 complex silence viral genomes? And how does the action of these complexes affect nuclear organization, gene expression, and genomic stability?
These are the kind of questions that keep us awake at night and drive our research. We are addressing such questions using a multi-disciplinary approach that involves genetics, genomics, biochemistry and imaging.
Read more about OUR RESEARCH.