Borsellini et al., Mol Cell, 2025
Switching on condensin
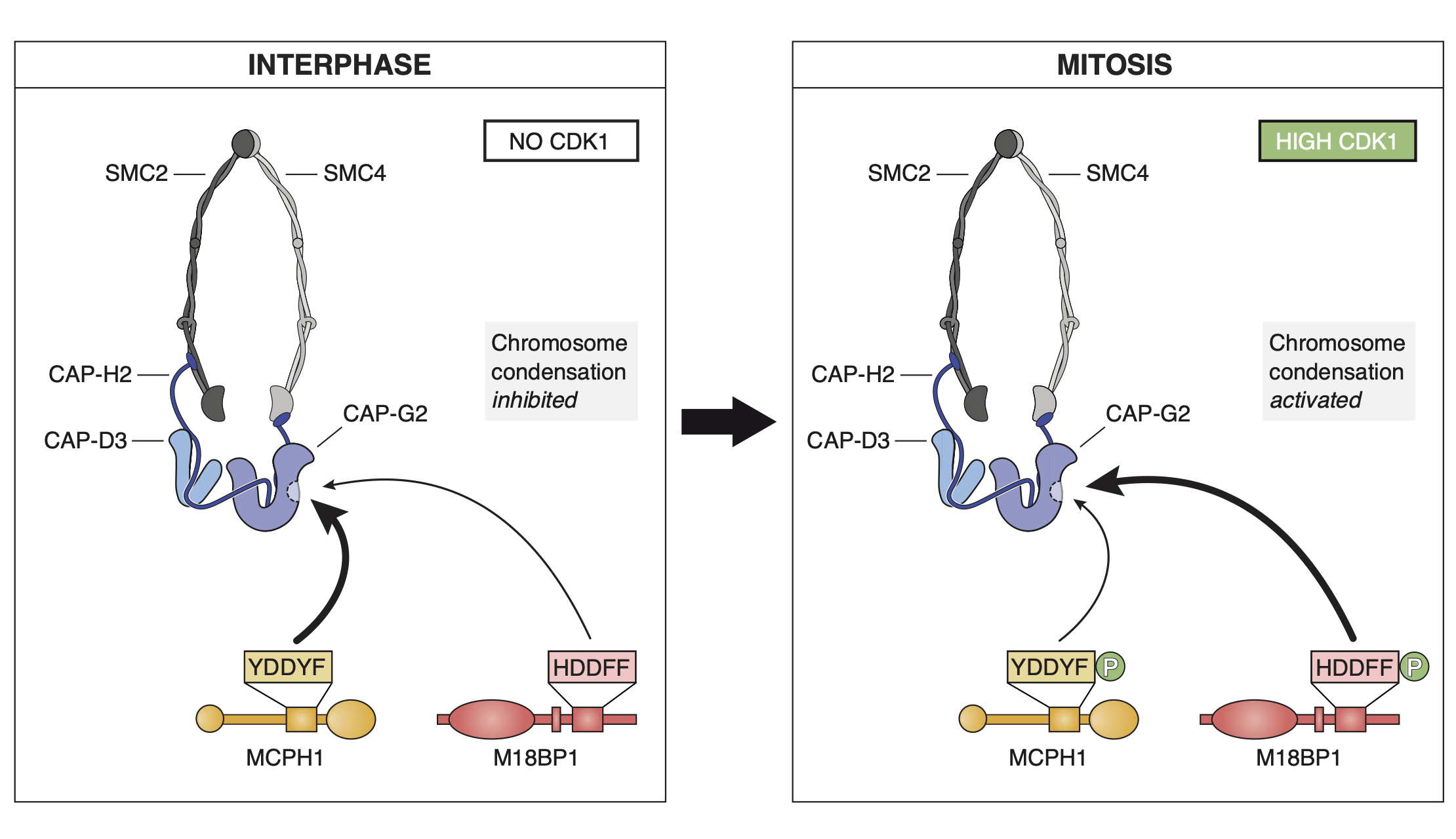
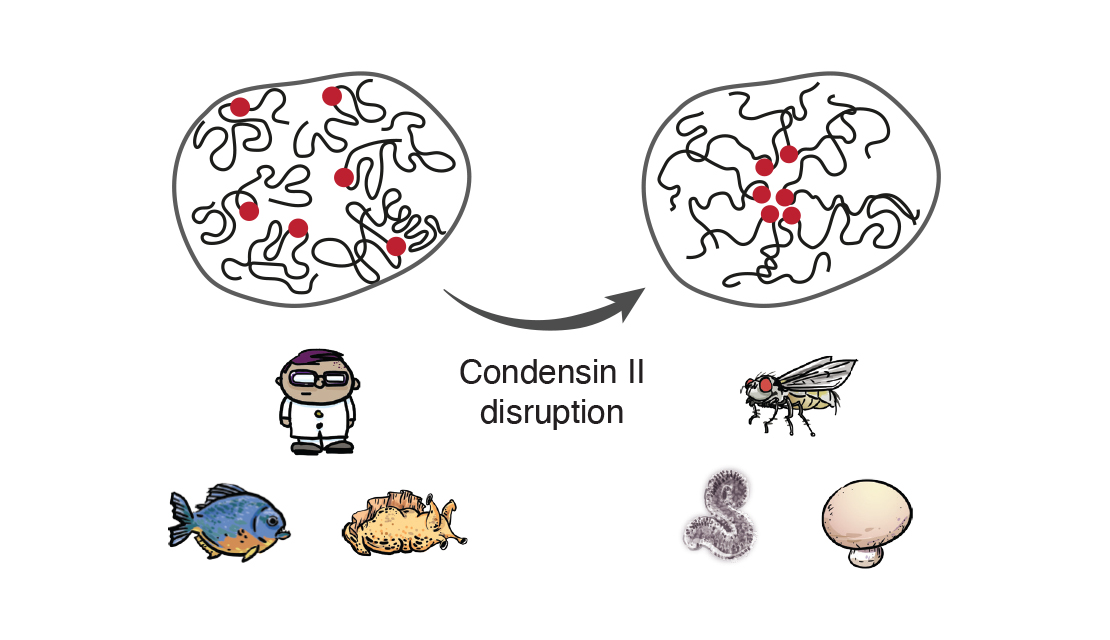
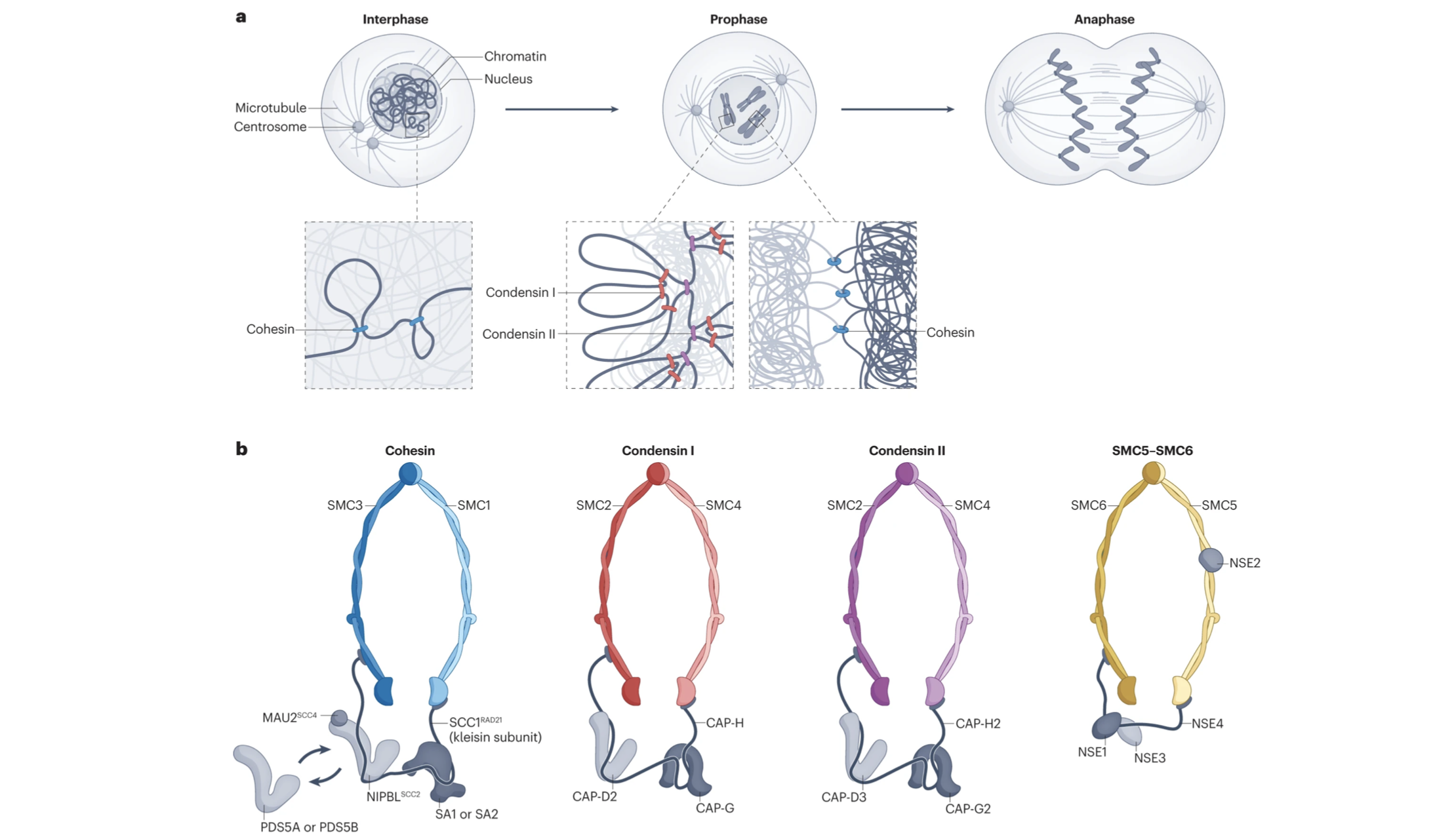
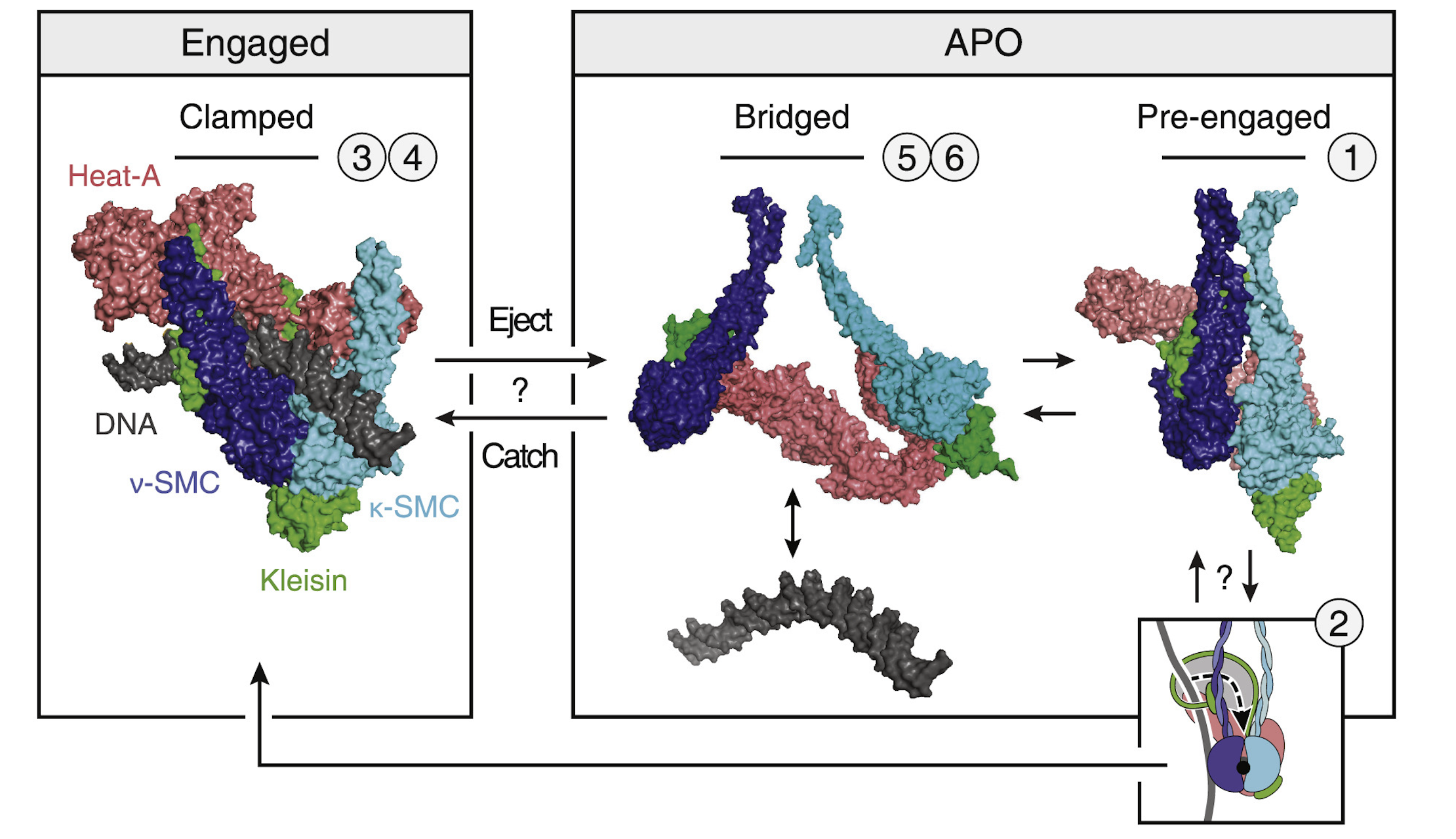
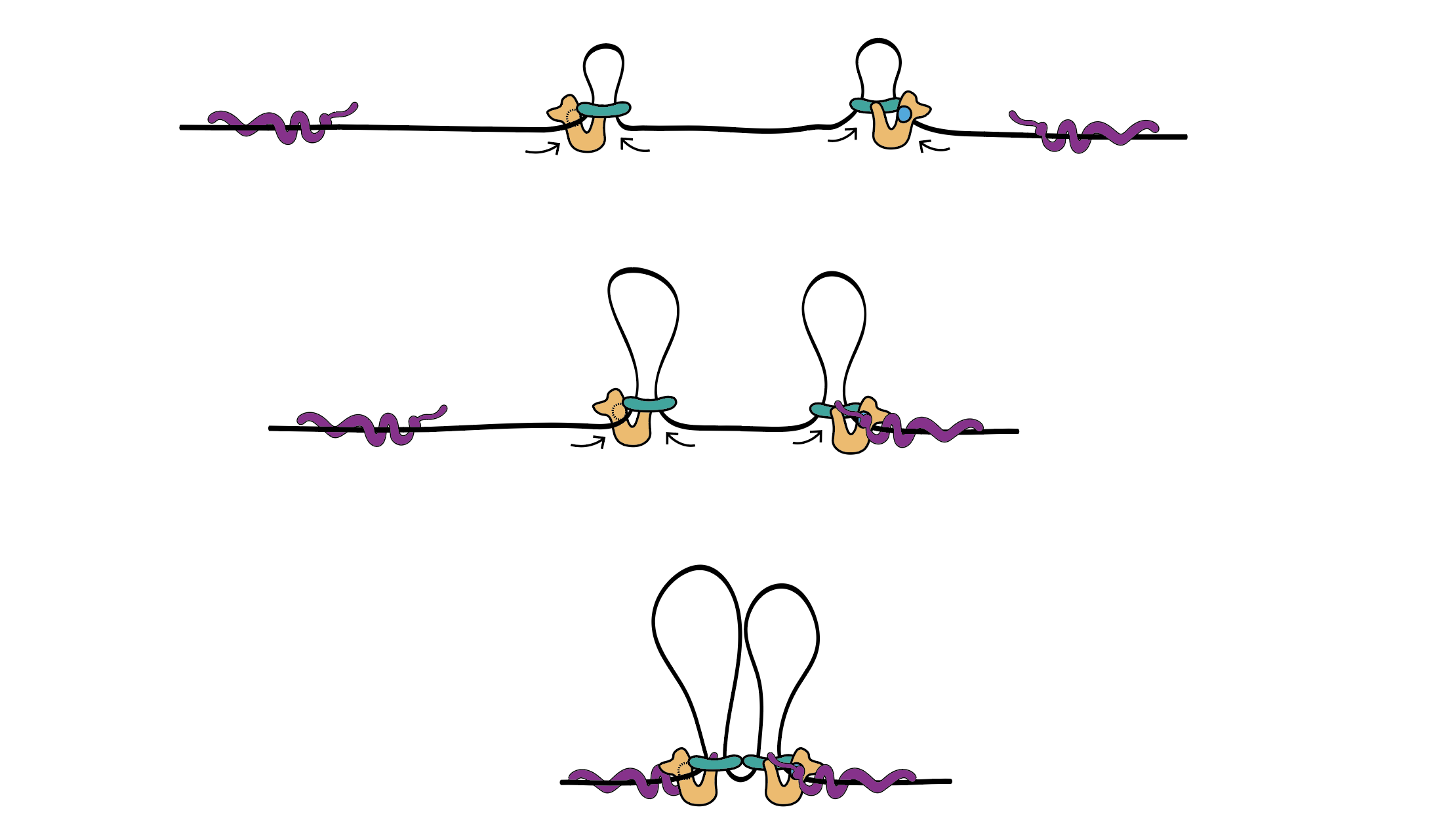
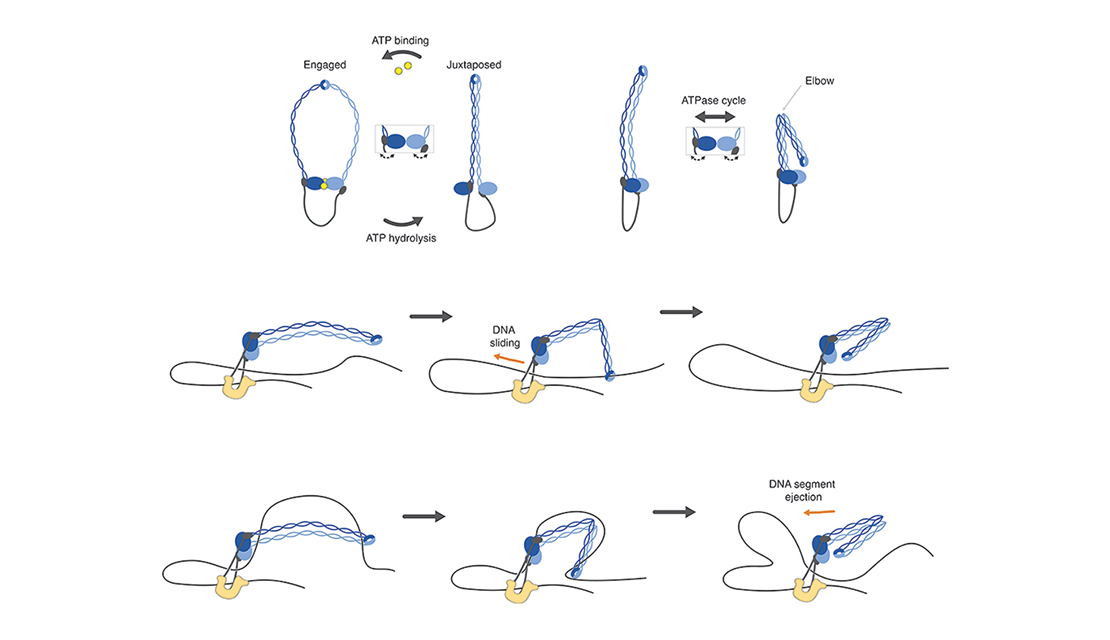
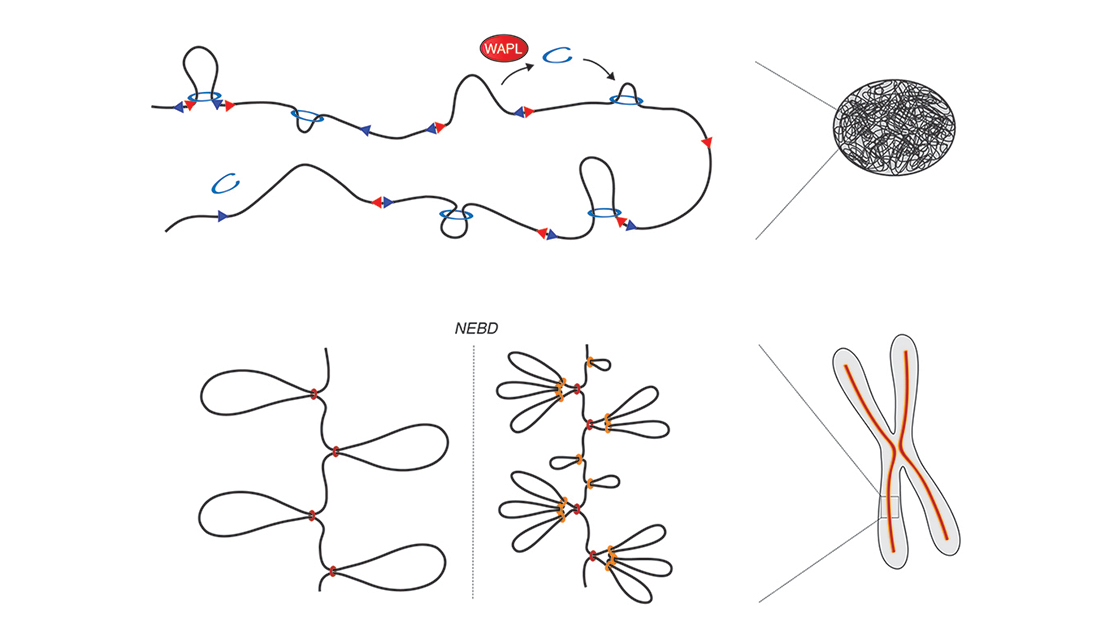
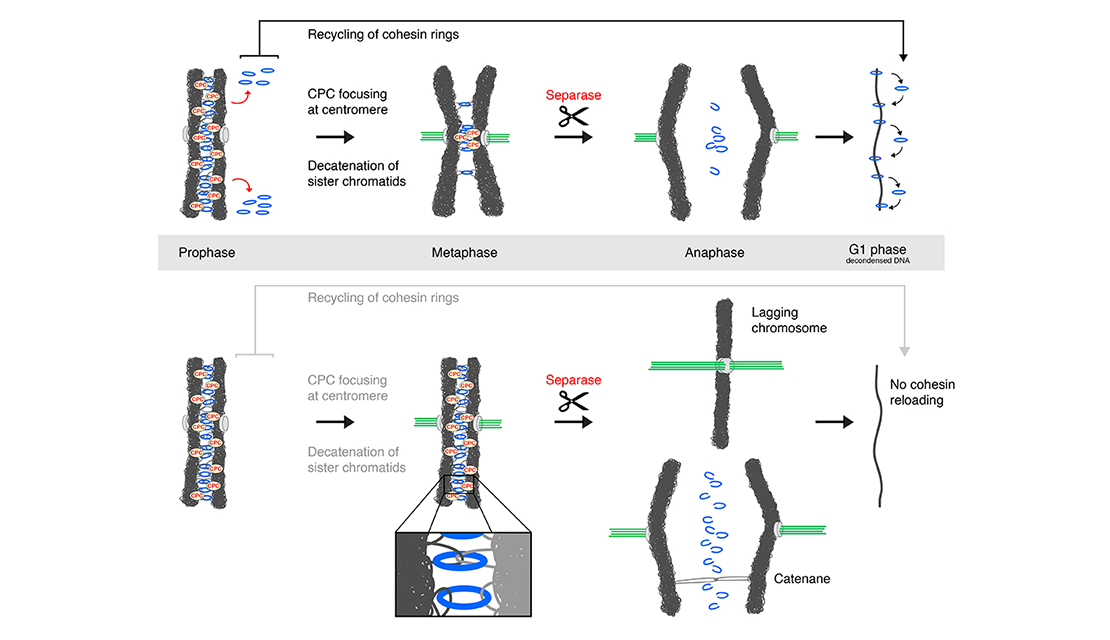
In a fabulous three-way collaboration with the Vannini and Musacchio labs, we discover what activates condensin as cells enter mitosis, and also what keeps the genome uncondensed during interphase. We find that M18BP1 binds condensin II to initiate condensation. MCPH1 keeps the interphase genome in its uncompacted state by outcompeting M18BP1 binding. The switch from MCPH1 to M18BP1 triggers chromosome condensation.
Check out OUR PAPER.