Our current research projects
These are the current projects in our lab. If you have any questions about the projects, do not hesitate to contact us.
Last update - 30-10-2025
Chromatin inside a cell nucleus
These are the current projects in our lab. If you have any questions about the projects, do not hesitate to contact us.
Last update - 30-10-2025
![OC Abstract Png 320[64]](/media/u3rihlxd/oc_abstract_png-320-64.jpg)
Promoters and enhancers are sensors and integrators of signals via transcription factors. Major questions about the interconnected function of promoters and enhancers are still unsolved: are there different types of promoters that depend or not in enhancer function? Which characteristics define each group of promoters? Do these play different roles in signal processing function or timing wise? How can we connect functionally promoters with their specific enhancer(s) in a systematic fashion? In this project, our goal is to develop an approach to deconvolute promoter autonomously driven responses from the enhancer-dependent ones; and stablish a method to rank/predict potential functional enhancers for enhancer-dependent promoters.
Contributor: Oscar Garcia Blay
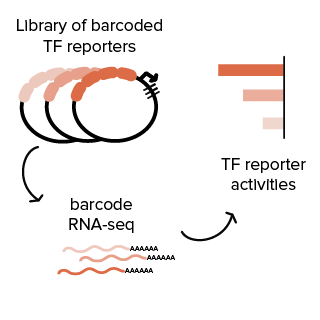
Transcription factors (TFs) respond to extra- and intracellular signaling cascades to collectively control the expression of the genome. To better understand how TFs mediate gene regulation, we aim in this project to directly measure the activity of many TFs in parallel using massively parallel reporter assays (MPRAs). Eventually, we also aim to integrate multiple TF reporters into the genome of cells in order to track the activity of multiple TFs in single cells in combination with single-cell mRNA sequencing. This technology platform of multiplexed TF reporters may be broadly used to study how cellular signaling events are transduced to the genome by TFs.
Contributors: Max Trauernicht and Hatice Yücel
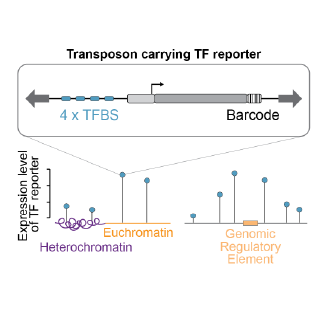
The activity of regulatory elements and transcription factors (TFs) depends strongly on their local molecular environment in the genome. In this project we are measuring the ability of individual TFs to activate transcription in thousands of genomic contexts in order to understand what determines their activity. We do this by combining two of our technologies: (1) TF reporter constructs that read out the activity of a single TF, and (2) transposon-based strategies for high-throughput insertion of sequences in random locations throughout the genome.
Contributor: Acadia Kocher
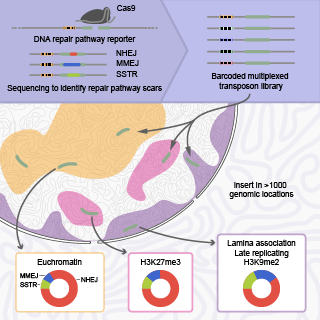
In this project we aim to understand how the balance between DNA repair pathways is affected by chromatin. In close collaboration with the Rene Medema lab we use our DSB TRIP pathway reporter in combination with genetic screens and small molecule screens to identify proteins that play a role in DNA repair in certain chromatin types.
Contributors: Xabier Vergara Ucin and Marcel de Haas
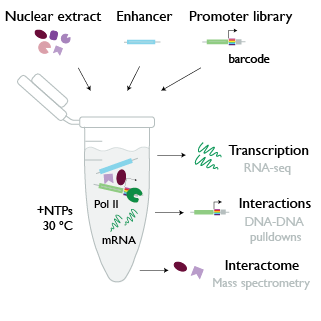
Developmental processes depend on distal enhancers that communicate with their target genes over large genomic distances in order to activate transcription. This project aims at investigating the biochemical mechanisms that enable enhancer-promoter communication and confer specificity. To this end, we establish enhancer-promoter complexes in vitro and read out transcriptional levels and interactions with a set of high-throughput assays. In collaboration with the Vermeulen lab, we further aim to identify protein interactors that are required for the information exchange at the enhancer-promoter pair.
Contributor: Pia Mach
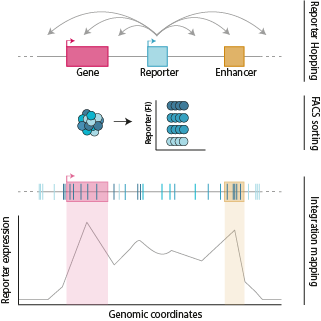
Genes are often activated by enhancers located at large genomic distances. The importance of this positioning on a linear scale is still poorly understood. To better understand positional relationships between regulatory elements, we developed a transposon-based technique that allows us to re-locate (‘hop’) almost any type of DNA to thousands of alternative genomic positions. By doing so we can generate high-resolution functional maps of genomic loci. We are currently investigating the positional relationship of a compatible promoter – enhancer pair but we anticipate that this technology can be used to study a wide range of biological questions.
Contributors: Mathias Eder and Christine Moene
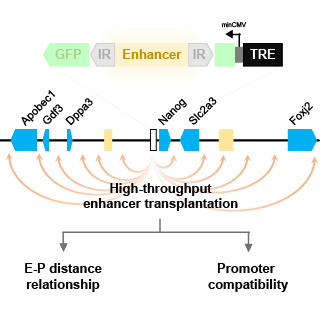
Enhancers are regulatory elements that can modulate the expression level of nearby genes through interactions with their promoter, and play an indispensable role in establishing the diverse gene expression patterns observed in different cell types. In this project, we use a transposon-based technique to translocate an enhancer into many different places in its native locus. By studying the consequences for local gene regulation, we hope to gain new insights about how local chromatin environments enable or constrain enhancer-mediated gene regulation.
Contributor: Martijn Verkuilen
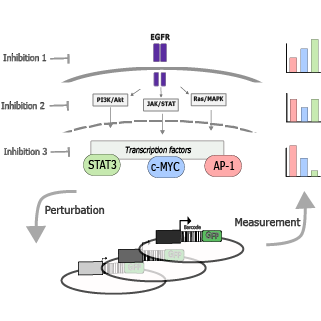
This project aims to decode the complex language of cancer cell signaling, focusing on the Epidermal Growth Factor Receptor (EGFR) pathway. By combining a novel multiplexed reporter system capable of tracking ~100 transcription factors simultaneously with advanced deep learning models, we're mapping out how EGFR signaling influences gene activity in cancer cells. Our goals are to identify the full range of transcription factors regulated by EGFR, determine their specific gene targets, and understand how different signaling branches (MAPK, PI3K/AKT, and JAK/STAT) contribute to this regulation.
Contributor: Daniel Goodall
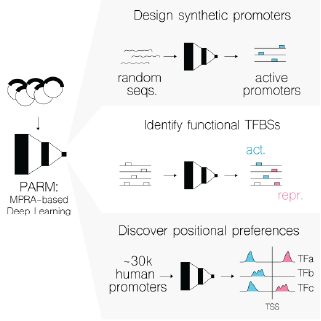
A key challenge in genomics is predicting gene expression from sequence alone, particularly promoters, whose regulatory logic remains unclear. Together with the lab of Jeroen de Ridder we developed PARM, a deep learning model that predicts promoter activity from DNA sequences, using much less computational power than existing methods. PARM is capable of designing synthetic promoters, can explain the cell-type specific regulatory logic of promoters through transcription factor regulatory sites and allows us to reveal positional preferences and motif interactions that differ between activators and repressors.
Contributors: Noud Klaassen, Vinicius Franceschini dos Santos, Marcel de Haas, Hatice Yücel and Lucia Barbadilla-Martinez (Jeroen de Ridder lab)
This website uses cookies to ensure you get the best experience on our website.