Judith Haarhuis
Associate staff scientist
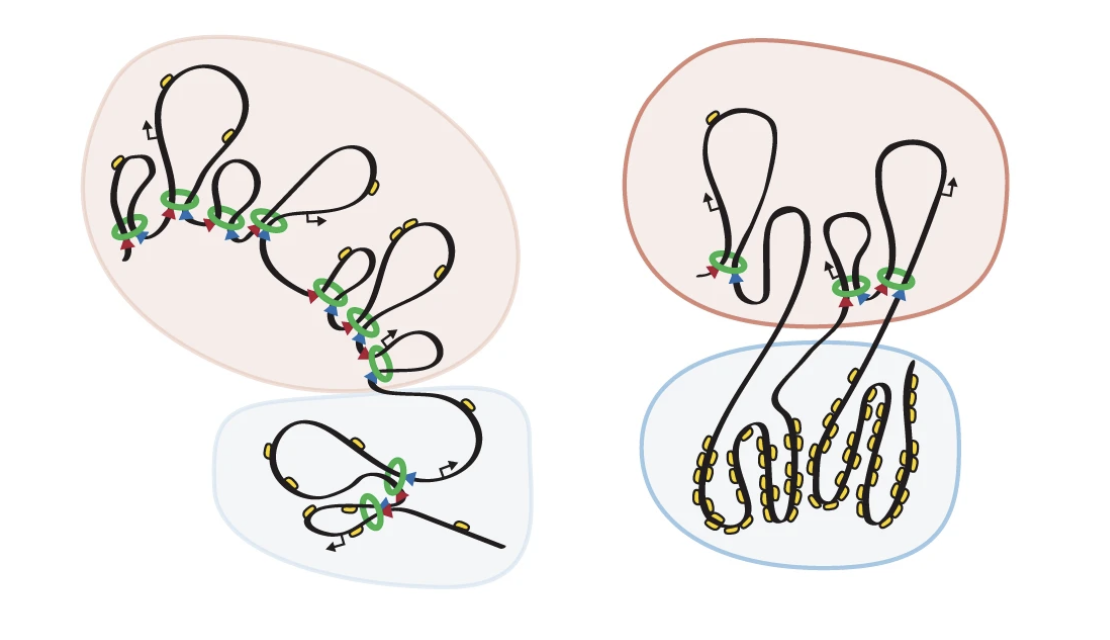
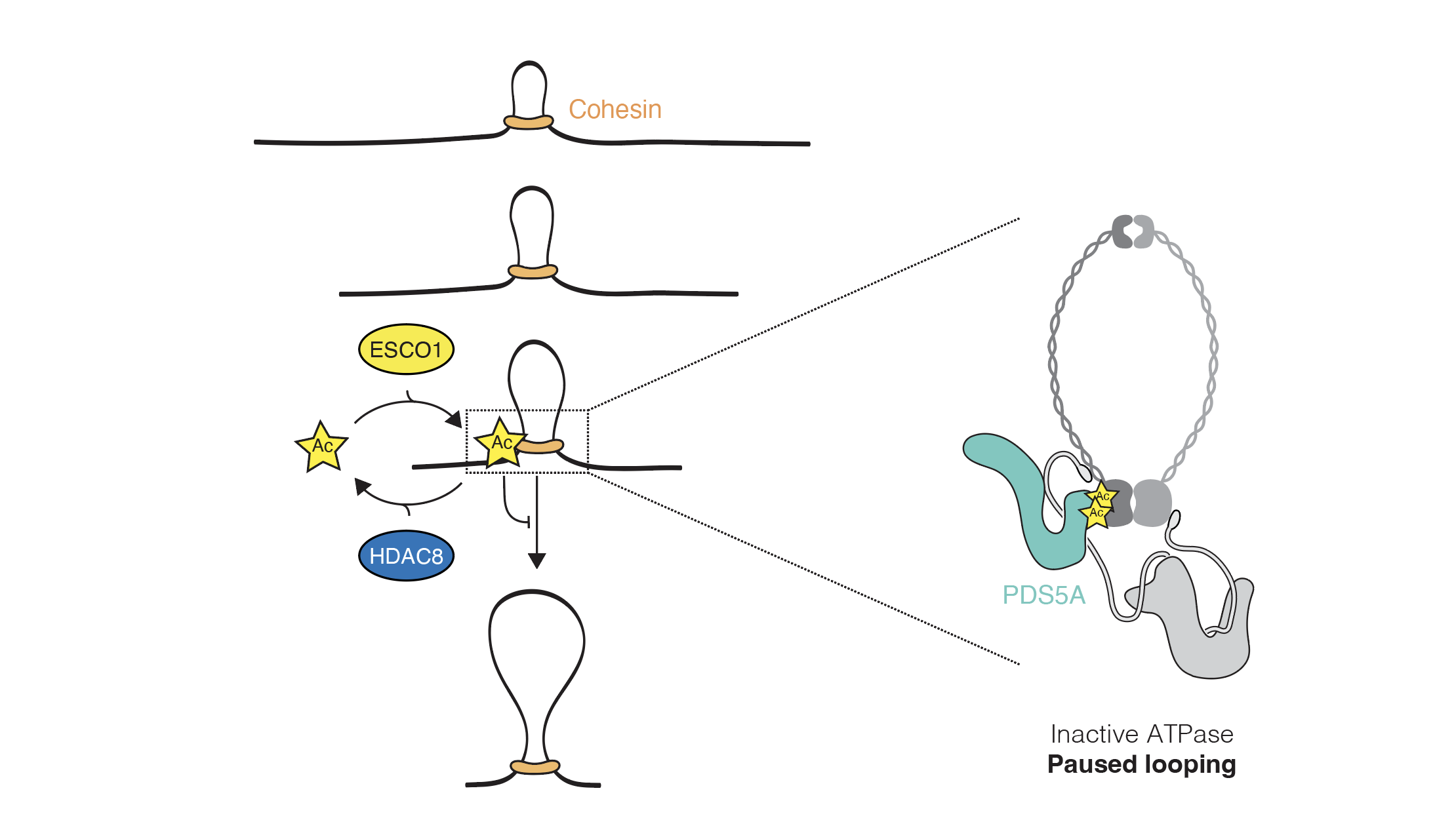
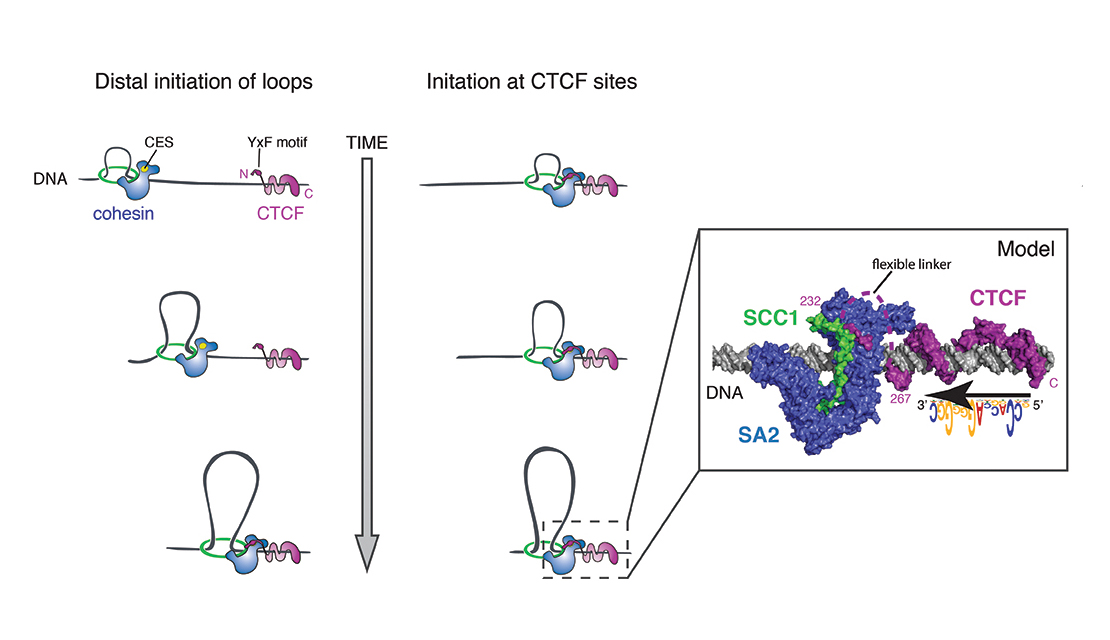
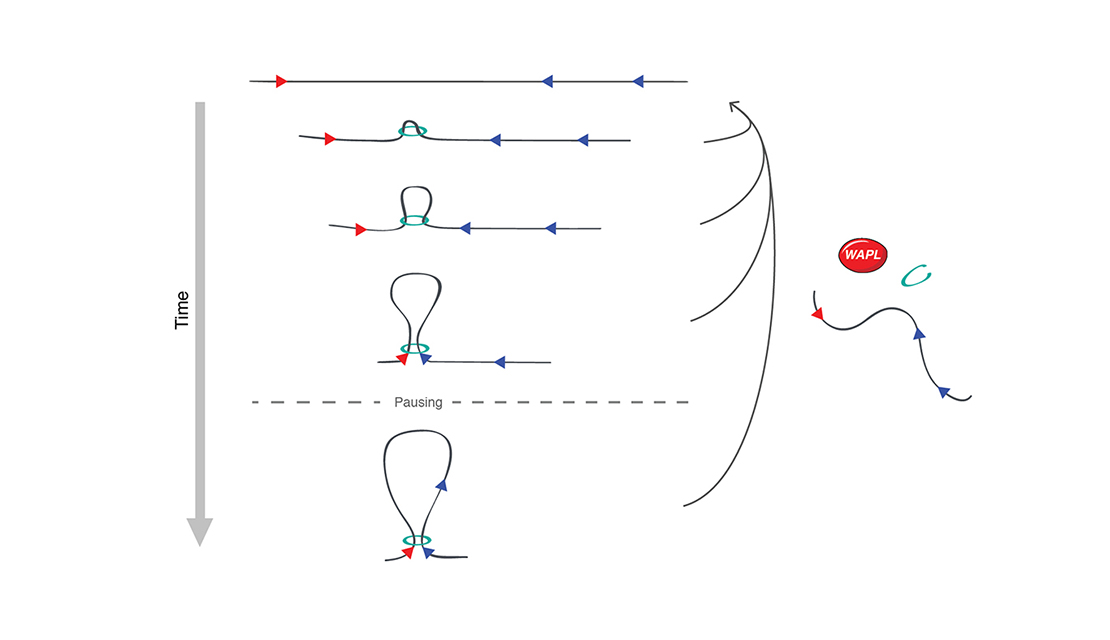
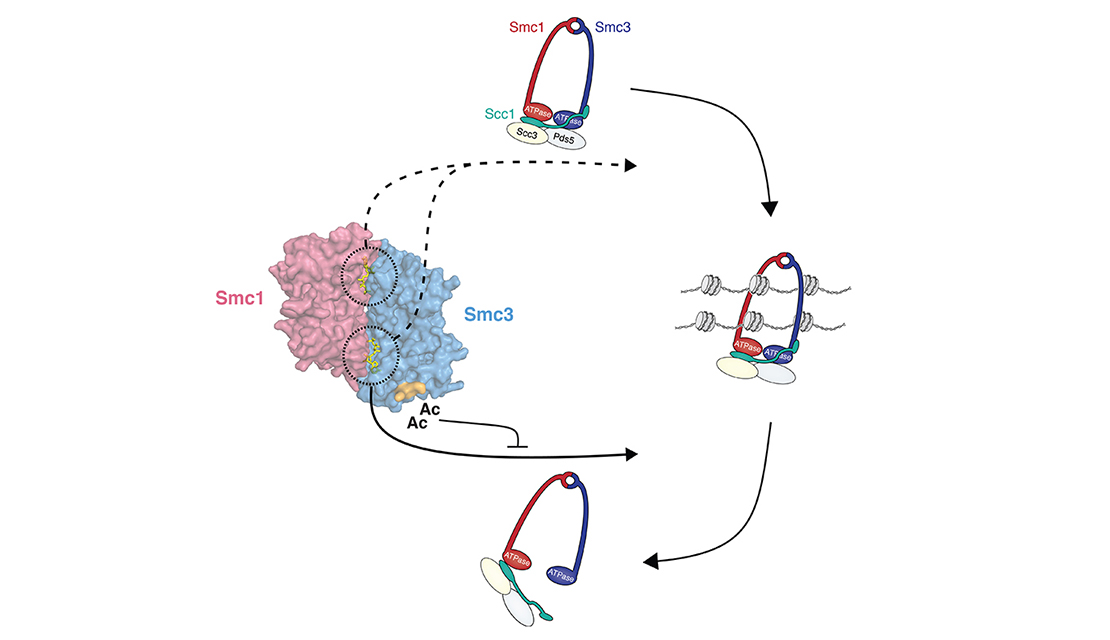
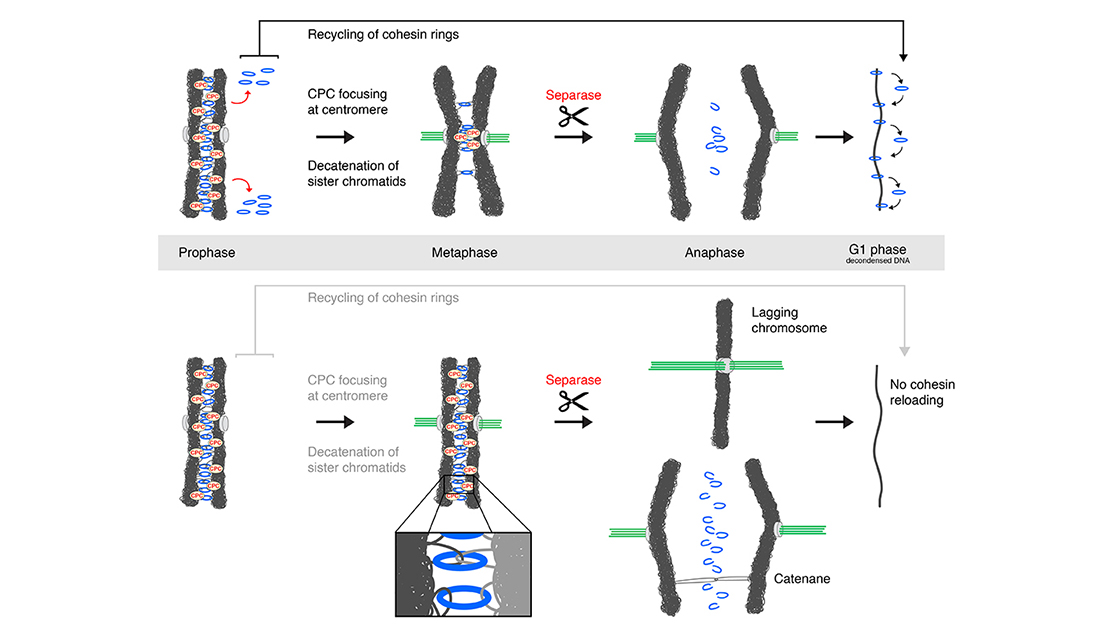
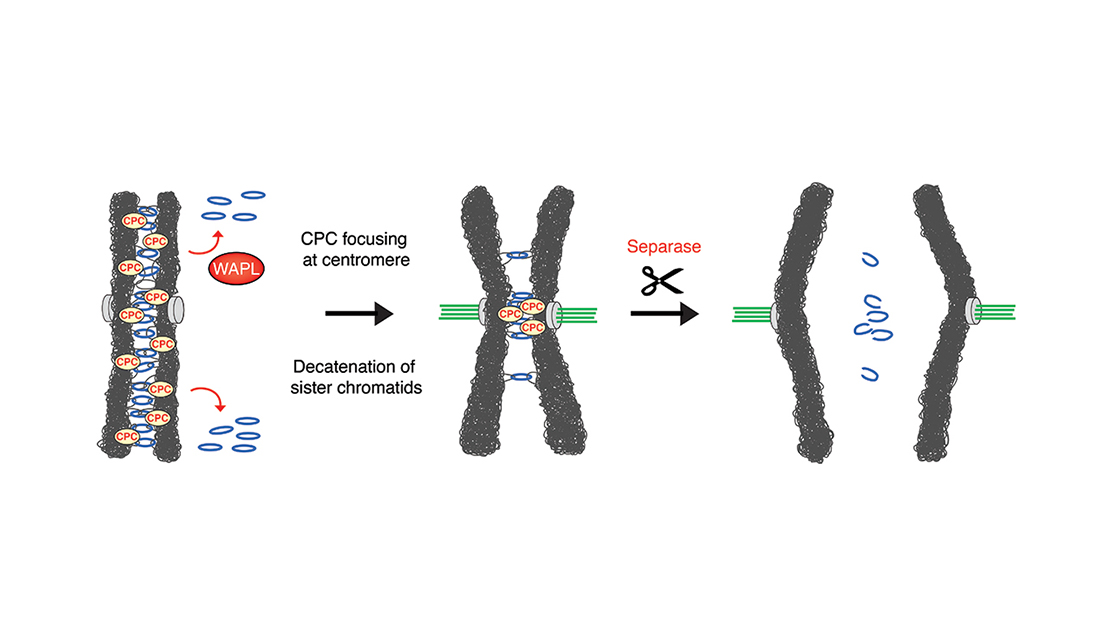
Email Benjamin RowlandMy interest lies in the molecular mechanism behind chromosomal architecture. DNA is structured via chromosomal loops created by SMC complexes. I studied how DNA loop extrusion by cohesin is regulated and what the effect of these loops is on cellular behaviour.
These days I focus on another SMC complex, called the SMC5/6 complex. SMC5/6 is involved in genomic maintenance, as it functions in DNA replication and DNA damage repair. Recently, it has been discovered that this complex can similarly to cohesin form DNA loops. Interestingly, the implications of these DNA loops, and the real mechanistic function whereby SMC5/6 acts, remain a mystery.