Champagne et al., Molecular Cell, 2021
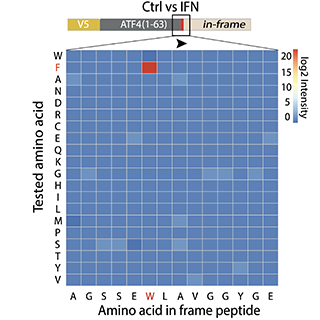
Tryptophan depletion results in tryptophan-to-phenylalanine substitutants
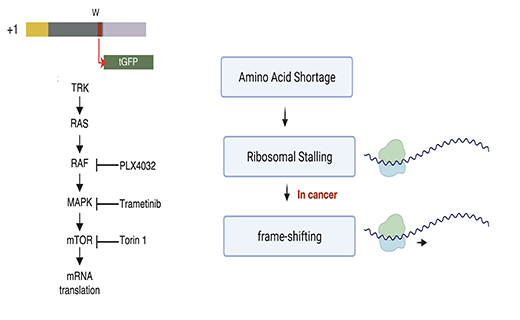
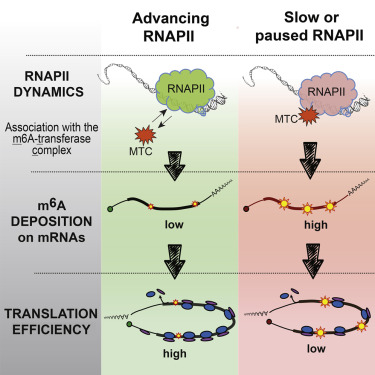
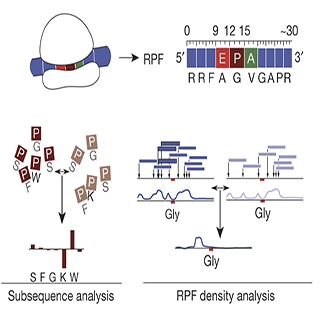
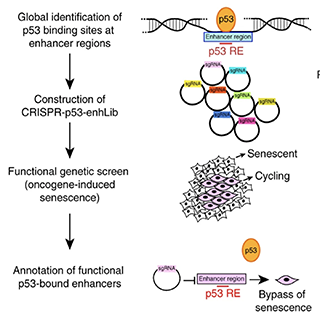
Activated T cells secrete interferon-γ, which triggers intracellular tryptophan shortage by upregulating the indoleamine 2,3-dioxygenase 1 (IDO1) enzyme. Such tryptophan shortage conditions stimulate the production of aberrant proteins by ribosomal frameshifting and codon reassignment events at tryptophan codons. We termed this phenomenon “sloppiness in mRNA translation” and strikingly observed its association with hyperactivation of the MAPK pathway. Functionally, sloppiness-induced events can impair protein activity, but also expand the landscape of antigens presented at the cell surface. These aberrant neoepitopes activate T cell responses, and thus can potentially be used to improve tumor immunoreactivity.
Read OUR PAPER