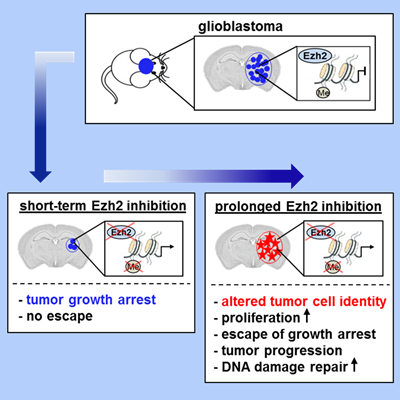
We recently demonstrated an oncogenic role for Ezh2 (histone methyltransferase and catalytic subunit of Polycomb repressive complex 2 (PRC2) in Kras driven non-small cell lung cancer. However, prolonged inactivation of PRC2 in aggressive Kras;P53 mutant NSCLC uncovered a profound tumor suppressive function for PRC2 loss resulting in tumor cell identity change, driven by inflammatory responses and EMT. This resulted in new vulnerabilities that can be exploited using combined inhibition of PRC2 and inflammatory responses. Ezh2 is overexpressed in glioblastoma multiforme (GBM) suggesting a possible oncogenic role. In a mouse model for GBM we demonstrated using inducible Ezh2 shRNAs and specific Ezh2 inhibitors that short-term intermitted inhibition indeed slowed tumor growth and prolonged survival. However, prolonged Ezh2 inhibition caused a robust switch in cell fate, resulting in enhanced proliferation and invasion, enhanced DNA repair and activation of a stem cell pluripotency network, resulting in therapy-resistant aggressive GBM. This illustrates that dosing of Ezh2 inhibition is critical, and Ezh2 inhibitors need to be used with caution. We are using these GBM models with CRISPR screens to find more effective combination therapies.