Changes in the chemical modifications of histones can lead to ‘epigenetic’ changes in gene expression and cancer. The Van Leeuwen lab studies mechanisms and principles of epigenetic regulation, using innovative proteomic, genetic, and (epi)genomics approaches. Our general strategy is to develop new tools and technologies, most recently two novel barcode-ChIP-sequencing approaches to discover epigenetic regulators and to decode the proteome of a single genomic locus. These innovations enable us to explore new areas of chromatin biology (e.g. histone dynamics and inheritance) and to dissect specific chromatin processes in high molecular detail (e.g. histone methylation). We take advantage of yeast as a powerful model system and in parallel we are developing tools in human cells to translate our findings to mammals. We use mouse models to unravel the role of chromatin-based mechanisms in T- and B-cell development in immunology and cancer.
Histone methylation from yeast to T cell differentiation and cancer
We previously discovered the yeast histone methyltransferase Dot1, which methylates lysine 79 of histone H3 (H3K79; Fig. 1) along gene bodies. This modifying enzyme influences gene regulation, DNA damage response, and oncogenic transformation in mammals. Despite these important functions, very little is known about the mechanisms of action and regulation of Dot1 in yeast and DOT1L in mammals. Using a combination of yeast genetics, DNA-barcode-based screens (Epi-ID), new methods to study histone dynamics (RITE), proteomics, and modeling we have obtained a comprehensive picture of regulation of Dot1/H3K79 methylation by a two-way crosstalk with histone H2B ubiquitination, co-factor metabolism, histone turnover, and histone deacetylation. The latter mechanism is conserved in mammals and is linked to lymphomagenesis in the mouse. Our recent studies show that DOT1L also plays a critical role in normal T and B cell differentiation. Our current research focusses on understanding how DOT1L affects T cell function and how this knowledge can be used to modulate the adaptive immune system.
Novel insights by novel technologies: decoding chromatin regulomes and proteomes by DNA barcode sequencing
Gene regulation involves interactions of specific genomic loci with many different proteins. How these interactions are orchestrated at any given location over time is largely unknown because systematically measuring chromatin modifications or protein-DNA interactions at a specific locus in the genome is challenging. Traditionally, reporter systems have been used but they often indirectly report on the chromatin status and therefore suffer from many possible biases. Direct capture-mass spectrometry approaches generally suffer from high background. To solve these problems, we developed DNA barcode-based Epigenetics technologies in yeast: Epi-ID (Fig. 2) and Epi-Decoder. Epi-ID (Fig. 3). Epi-ID is aimed at identifying in a genome-wide manner regulators of known chromatin marks or chromatin-binding proteins Epi-Decoder is very powerful, versatile, and robust strategy to identify and quantify in an unbiased and systematic manner the proteome of an individual genomic locus by DNA sequencing. Epi-Decoder thus offers a unique approach to unravel protein-based mechanisms of gene regulation. Our current studies take advantage of CRISPR/Cas9 based genome editing and Epi-Decoder to discover novel chromatin proteins and novel mechanisms of regulation, with a special interest in H3K79 methylation and other chromatin modifications in gene bodies. In parallel we are developing strategies for barcode-based epigenetics screens in mammalian cells.
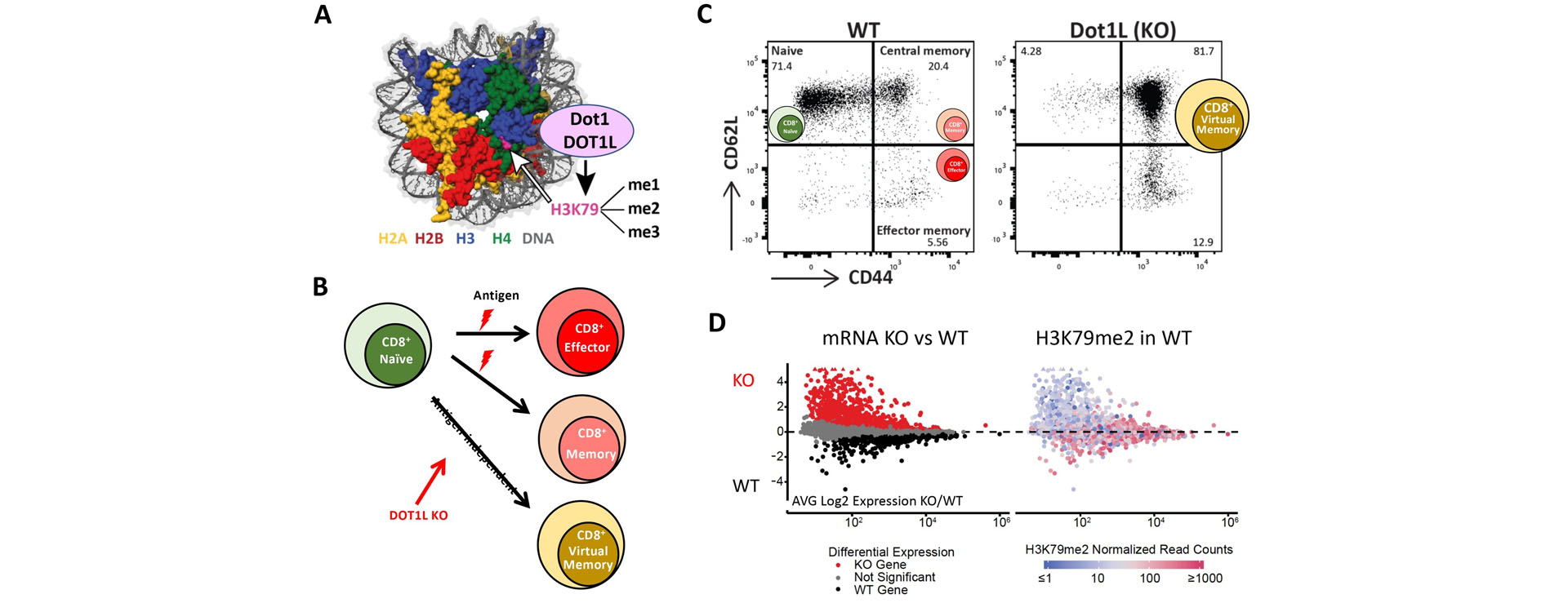
Figure 1. Dot1/DOT1L methylates H3K79. Upon deletion of Dot1L in the T-cell lineage (KO), cytotoxic (CD8+) T cells differentiate in an antigen independent manner to ‘virtual memory’ T cells. In KO vs WT memory-phenotype cells, many genes are derepressed and some genes show reduced expression. The latter category of genes is enriched for H3K79me2 in their gene bodies, while the derepressed genes lack H3K79me2 and are hence expected to be affected by indirect mechanisms.
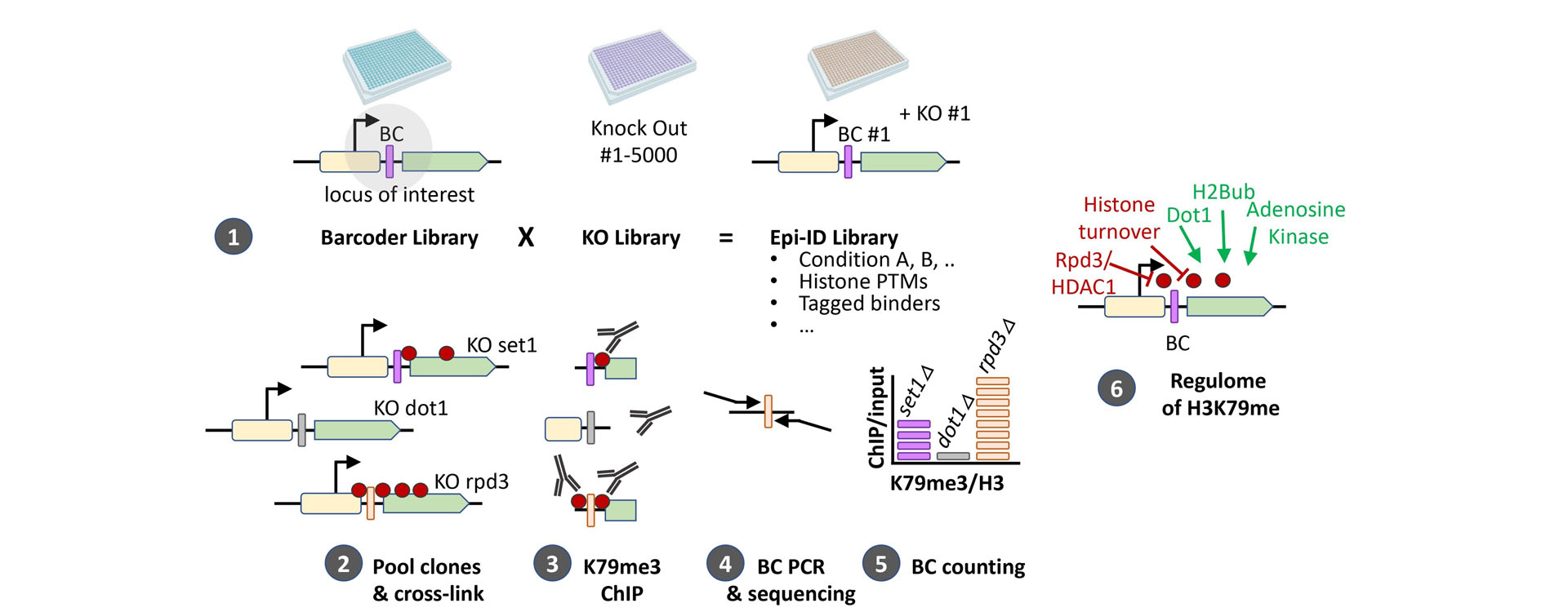
Figure 2. Epi-ID systematically determines which genes affect the abundance of a factor or interest at a barcoded (BC) locus of interest (e.g. histone modification, transcription factor, novel chromatin factor). Epi-ID involves combining a genome-wide library of knock-out strains with a library in which each yeast strain contains one or a pair of unique barcodes at a common locus. Using ChIP for the chromatin mark of interest, here H3K79me3, followed by barcode amplification, sequencing, and counting, we obtain a H3K79me3 ChIP/input value for each knock-out at the barcoded locus.
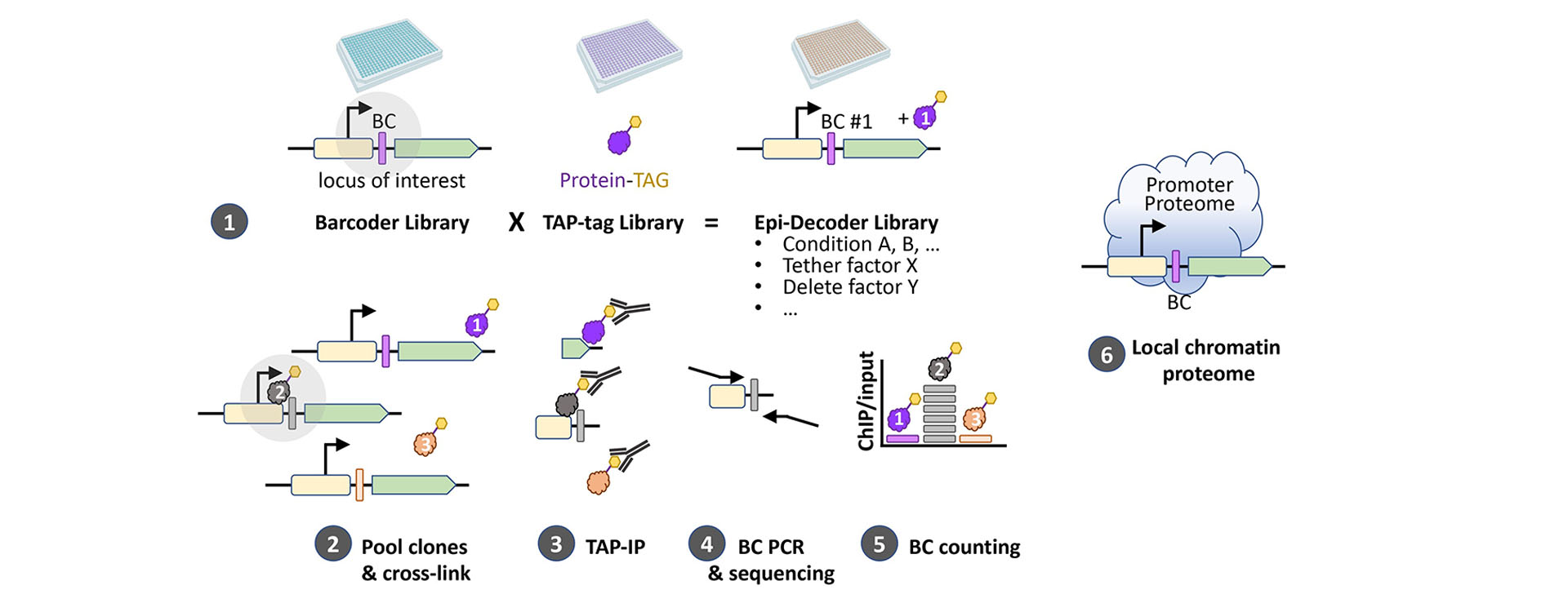
Figure 3. Epi-Decoder defines the local proteome around a barcoded (BC) locus of interest. Epi-Decoder involves combining a genome-wide library of epitope-tagged strains with a library in which each yeast strain contains one or a pair of unique barcodes at a common locus. Using tag-ChIP followed by barcode amplification, sequencing, and counting, we obtain a ChIP/input value for each protein at the barcoded locus. Epi-Decoder is versatile, and robust strategy to identify and quantify in an unbiased and systematic manner the proteome of an individual genomic locus by DNA sequencing without the need for extensive purification and mass spectrometry.